


The dBlast package is a wrapper for distributed NCBI BLAST[1], and was designed to run BLAST on Beowulf clusters, SMP machines, and MOSIX machines, without the need to buy/build/use a parallel BLAST version (MPI/PVM...). dBlast uses all of the normal NCBI BLAST code, and adds two extra command-line flags. The advantages by doing so:
Download the Manual(ASCII/PDF/PS)
dBlast - Documentation - last updated 15 April 2003
dBlast - A wrapper to run NCBI BLAST in parallel/distributed - Version 1.1 on Beowulf clusters, SMP machines, and MOSIX machines. Copyright © 2003 - Marc A. van Driel, Maarten L. Hekkelman, CMBI - All Rights Reserved.
The Basic Local Alignment Search Tool (BLAST) was designed to perform a rapid sequence comparison, generate (approximated) alignments of local similarity [1]. BLAST is a widely used program nowadays and thereby two type of problems can occur when A. a lot of people want to perform lots of BLAST jobs or B. very big jobs. There are a number of solutions to this problem:
Stop reading if you prefer option 1, otherwise it might be interesting for you. The second option is limited in its scaling and as the databases are still rapidly growing, this may be not the preferable option to choose. If you want to spend your money on commercial solutions, than that is up to you. The dBlast package was designed for options 4 and 5.
ad. option 4:
The BLAST algoritme is capable to run on SMP machines by using multi-threading (the -a option). However, this is limited in scaling and unfortunately does not use all the available cpu rescources. This effect can be seen on machines with more than 4 cpus.
ad. option 5:
For the first "problem", that is "to perform lots of BLAST jobs", most people use nowadays the queuing system to submit the jobs to the cluster (e.g. via PBS). However for the second "problem", that is very big jobs (or you just want to have a quicker response), this is not trivial. There are commercial BLAST implementations which use MPI (message passing interface) to run the BLAST job in a cluster environment. Unfortunately you have to buy them (at least for the moment) and more importantly, they are not maintained and updated by the NCBI. Furthermore, not all the MPI BLAST versions are producing the same values as a single BLAST run (e.g. E-scores). There is another disadvantage of using a MPI BLAST: the databases are read at one central place. Although this looks to be an advantage, and from a maintainers view it is, this can become slow because of the network traffic.
An solution to the "very big jobs (or you just want to have a quicker response)" problem is to give each processor/node a part of the total job. On the WWW there are tools available to split your database files and to merge the output of the BLAST "sub-jobs" afterwards. So what does dBlast extra? Nothing.. only (if we may say so, better) it is a ready to use package to do the job and (most important) the output is the same as you would get from a single run.
In contrast to the majority of tools, dBlast redivides the database files not only equally, but also in a balanced way. This gives you a balanced load foreach processor/node/job (more in chapter 3). The merge phase is also different than most tools. The BLAST algorithm is continuesly updating and the statistics are depending on the query length, database size, the BLAST algorithm chosen (blastn, blastx...), the scoring matrix specified by the user, and most of the other options given by the user. So, a simple text merge of the "sub-job" will result in a faulty output. Using the -z option to specify an effective database size is not enough to correct this, the reason for this is that the effective database size AND the effective query length are needed and both are calculated using the length adjustment which are normally not available when you launch blastall for only a part of the total database [2]. In the dBlast package this problem is tackled by a patch for the NCBI BLAST. By applying this patch, you can get these critical (statistical) values, based on all the options specified by the user. Subsequently, the "sub-jobs" are started with these values and the "sub-jobs" will have the same statistics as if it were one BLAST job. In the end a simple text merge will do the final touch.
More background on specific parts of dBlast can be found at the appropiate site in this file.
The dBlast package contains the following programs/scripts:
The following packages are required. The tested versions are placed between parenthesis.
The package can be unpacked at any place on your system. It is however necessary to have the patched BLAST (and the data directory containing the matrices,..see below) and dmerge in the same directory.
The package can be unpacked at any place on your system, as long as the directory is shared between the nodes of the cluster. The patched BLAST (and the data directory containing the matrices,..see below) and dmerge should be in the same directory.
After downloading and uncompressing the NCBI toolkit, which can be found at the NCBI ftp site:
ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools/ncbi.tar.gzthe patch (ncbi.patch) can be applied in the following way:
patch -p0 < ncbi.patch
(in the same directory as where the ncbi directory is located) The NCBI toolkit can now be build as described in it documentation. The blastall executable can be found in 'ncbi/build/'. After compilation be sure that you copy either the 'blastall' executable together with the data directory containing the BLAST matrices (BLOSUM62...), in the same directory as 'dmerge' (or the other way a round).
To test if the patch and compilation of blast worked, you can execute 'blastall' and the extra options (see below) should appear at the bottom of the options list.
-B Print Length Adjustment and quit [T/F]
default = F
-C Length Adjustment to use [Real]
default = 0
The variables used:
| Name | Explaination (example value) |
|---|---|
| DBLAST_THREADS | The number of threads used by dformatdb (2 in a dual processornode) |
| DBLAST_BLASTALL | The patched blastall executable (/usr/local/blast/blastall) |
| DBLAST_MACHINETYPE | The machine type you are running dBlast on (CLUSTER or MOSIX/SMP) |
| DBLAST_NODES | Number of nodes/processors to BLAST on (24: number of processors, e.g. 12 dual processor machines; this value is also used to divide the database(s) in parts) |
| DBLAST_FORMATDB | The formatdb executable (/usr/local/blast/formatdb) |
| DBLAST_DMERGE | The dmerge executable (/usr/local/blast/dmerge) |
Before you continue, please set the environment variables:
| SMP/MOSIX (24 processor machine) |
Cluster (Cluster with 12 dual processor nodes) |
|---|---|
| in bash/sh | |
| export DBLAST_THREADS=32 | export DBLAST_THREADS=2 |
| export DBLAST_BLASTALL=/usr/local/blast/blastall | export DBLAST_BLASTALL=/usr/local/blast/blastall |
| export DBLAST_MACHINETYPE=SMP (or MOSIX) | export DBLAST_MACHINETYPE=CLUSTER |
| export DBLAST_NODES=24 | export DBLAST_NODES=24 |
| export DBLAST_FORMATDB=/usr/local/blast/formatdb | export DBLAST_FORMATDB=/usr/local/blast/formatdb |
| export DBLAST_DMERGE=/usr/local/blast/dmerge | export DBLAST_DMERGE=/usr/local/blast/dmerge |
| in tcsh/csh | |
| setenv DBLAST_THREADS 32 | setenv DBLAST_THREADS 2 |
| setenv DBLAST_BLASTALL /usr/local/blast/blastall | setenv DBLAST_BLASTALL /usr/local/blast/blastall |
| setenv DBLAST_MACHINETYPE SMP | setenv DBLAST_MACHINETYPE CLUSTER |
| setenv DBLAST_NODES 24 | setenv DBLAST_NODES 24 |
| setenv DBLAST_FORMATDB /usr/local/blast/formatdb | setenv DBLAST_FORMATDB /usr/local/blast/formatdb |
| setenv DBLAST_DMERGE /usr/local/blast/dmerge | setenv DBLAST_DMERGE /usr/local/blast/dmerge |
dBlast redivides the database files not only equally, but also in a balanced
way. This will give you a balanced load foreach processor/node/job. The FastA
formatted database can be found at several ftp-sites around the world. To
redivide a database you can take one or more files as input for the dformatdb
program. The dformatdb program is a wrapper for formatdb. It takes the normal
formatdb options, but uses the DBLAST_NODES environment variable to divide the
database(s) in the number specified. dformatdb is a multitreaded application
and uses DBLAST_THREADS to find the number of threads. Be sure that the
DBLAST_FORMATDB environment variable is set to the executable formatdb (e.g.:
/usr/local/blast/formatdb). dformatdb -h gives you all the
options:
dformatdb [options] -n outFileBaseName file [files...]
-p[T/F] Type of file, T = protein (default T)
-o[T/F] Parse SeqId and create indexes (default F)
-s[T/F] Create indexes limited only to accessions - sparse (default F)
-A[T/F] Create ASN.1 structured deflines (default F)
-l logfile logfile name (default formatdb.log)
An example:
bash$> export DBLAST_NODES=24 bash$> export DBLAST_THREADS=32 bash$> export DBLAST_FORMATDB=/usr/local/blast/formatdb bash$> dformatdb -pT -oT -sT -n sprot /data/download/fasta/sprot.fa
The result will be sprot00, sprot01, ...., sprot23 blast index files.
NOTE: The redived FastA files are not generated. If you want to save the FastA files, you can change 'DEFINES = PIPE_TO_FORMATDB=1' to zero.
The idea of d(istributed)blast is to distribute the databases to the computing nodes. That is on the local disks. The main reason for this is that the NFS setup is slow, and that the PVFS setup is (unfortunately) not always stable. If you want to run on a cluster you can create a directory (e.g. /data) on the local disk of every node and copy the formatted databases (3.2) to this directory. We usually use 'scp' to do so.
An example to distribute the databases to the nodes:
for i in 02 03 04 05 06 07; do scp mgr@cluster:/data/sprot??.*
mgr@node$i:/data;done
You can of course use your own way to distribute the files using e.g. rsync.
dBlast can be started by the dblastall.pl script. The script takes almost the same parameters as blastall does, except the following:
-z Effective length of the database (use zero for the real size) [Real]
default = 0
-a Number of processors to use [Integer]
default = 1
The effective length is calculated by the dblastall.pl script before running blastall and the number of processors is determed by (environment variable) DBLAST_NODES
The patched options in NCBI blast:
-B Print Length Adjustment and quit [T/F]
default = F
-C Length Adjustment to use [Integer]
default = 0
Before you run dBlast, please set DBLAST_BLASTALL, DBLAST_MACHINETYPE, and DBLAST_DMERGE.
An example to use dBlast:
dblastall.pl -p blastp -d /data/sprot -i /home/mgr/blastp.seq
THE DBLAST PACKAGE LICENSE INFORMATION: COPYRIGHT © 2003 BY MARC A. VAN DRIEL AND MAARTEN L. HEKKELMAN ALL RIGHTS RESERVED. THIS SOFTWARE (THE DBLAST PACKAGE) IS FURNISHED UNDER A LICENSE AND MAY BE USED AND COPIED ONLY IN ACCORDANCE WITH THE TERMS OF SUCH LICENSE AND WITH THE INCLUSION OF THE ABOVE COPYRIGHT NOTICE. THIS SOFTWARE OR ANY OTHER COPIES THEREOF MAY NOT BE PROVIDED OR OTHERWISE MADE AVAILABLE TO ANY OTHER PERSON. NO TITLE TO AND OWNERSHIP OF THE SOFTWARE IS HEREBY TRANSFERRED. THE INFORMATION IN THIS SOFTWARE IS SUBJECT TO CHANGE WITHOUT NOTICE AND SHOULD NOT BE CONSTRUED AS A COMMITMENT BY THE CMBI OR BY THE UNIVERSITY OF NIJMEGEN, OR BY THE AUTHORS. NEITHER THE CMBI, NOR THE UNIVERSITY OF NIJMEGEN NOR THE AUTHORS ASSUME RESPONSIBILITY FOR THE USE OR RELIABILITY OF THIS SOFTWARE PRODUCT. IT IS NOT ALLOWED TO REDISTRIBUTE THE (SOURCE) CODE. ALSO NOT ALLOWED IS THE USAGE IN OTHER PROGRAMS. IT IS NOT ALLOWED TO MODIFY THE CODE. DBLAST IS WRITTEN BY MARC A. VAN DRIEL AND MAARTEN L. HEKKELMAN FOR FREE USAGE BY ACADEMIC AND OTHER NON-PROFIT ORGANIZATIONS. RESULTS OBTAINED BY THIS PROGRAM CAN FREELY BE PUBLISHED PROVIDED THE PROGRAM AND ITS AUTHOR ARE ACKNOWLEDGED BY NAME. COMMERCIAL ORGANISATIONS CAN OBTAIN A LICENSE TO USE DBLAST SUCH A LICENSE GIVES THEM THE FULL RIGHTS TO DO EVERYTHING WITH IT THEY WANT, EXCEPT REDISTRIBUTE, OR PUBLISH THE PROGRAM.
[1] Altschul, Stephen F., Gish Warren, Webb Miller, Eugene W. Myers, and David J. Lipman (1990). Basic local alignment search tool. J. Mol. Biol. 215:403-410.
[2] Altschul, Stephen F., Gish Warren (1996). Local alignment statistics. Meth. Enzymol. 266:460-480.
Version 1.0 (25-11-2002) INITIAL RELEASE
Version 1.1 (13-04-2003) Public release
Version 1.1.1 (05-06-2003) Patch update for NCBI Blast version 2.2.6
Please contact us to report bugs and other remarks. If you are unable to install dBlast, please read the manual first.
Centre for Molecular and Biomolecular InformaticsNote: Please read the license information before you download the program/code.
| Item | HTTP | FTP |
|---|---|---|
| Manual | ASCII | ASCII |
| PS | PS | |
| Source code | Source | Source |
| Precompiled binaries | Linux x86 | Linux x86 |
| IRIX 32 | IRIX 32 | |
| FreeBSD | FreeBSD |
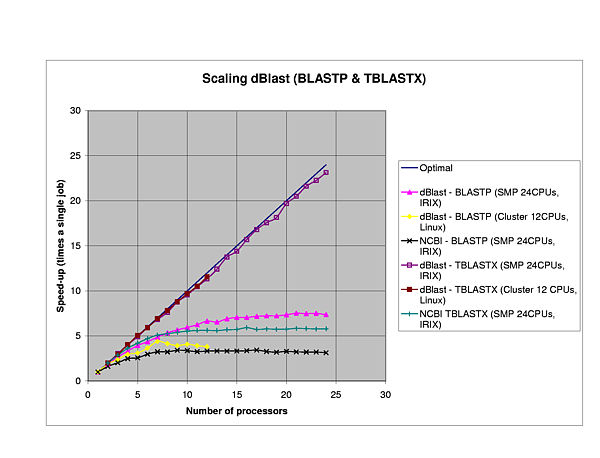
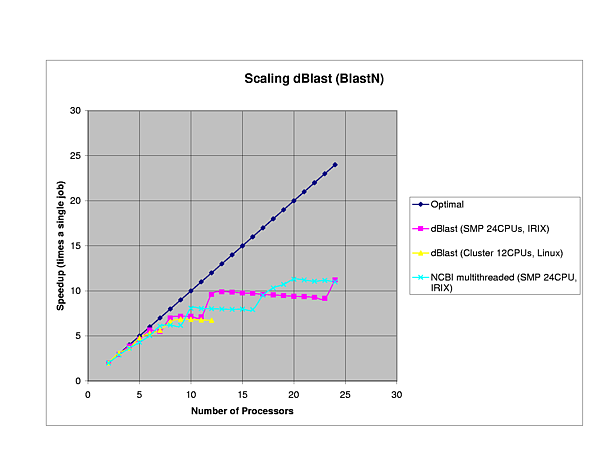
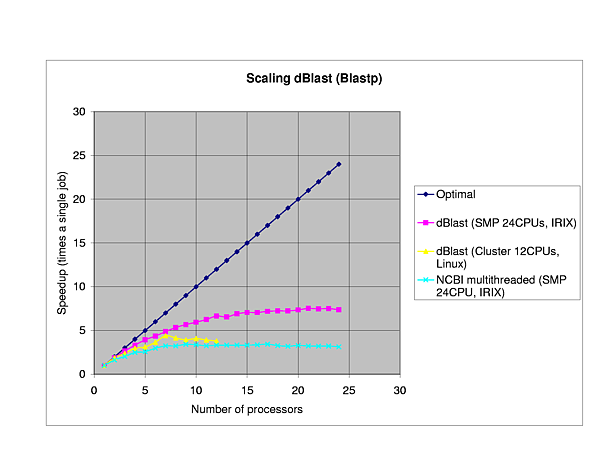
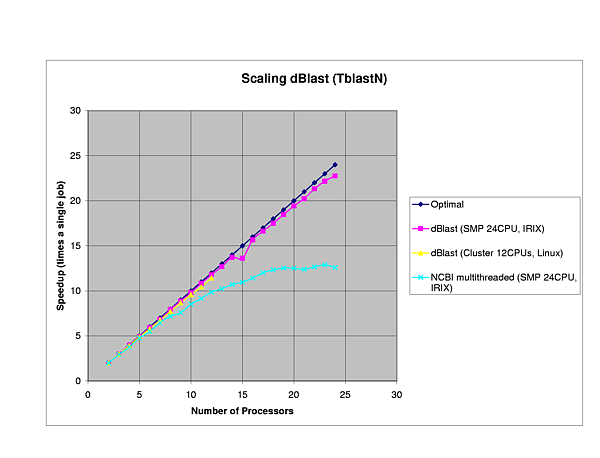
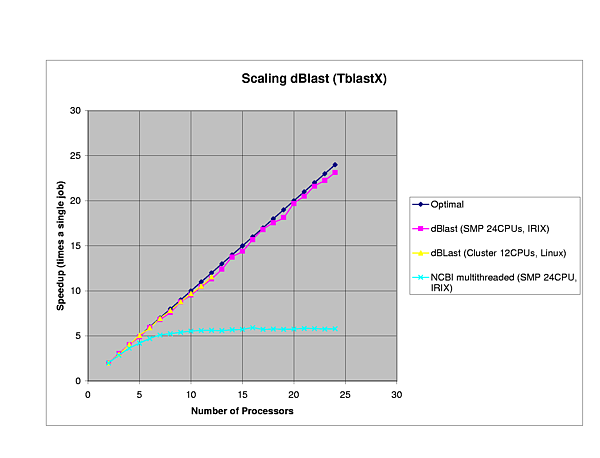
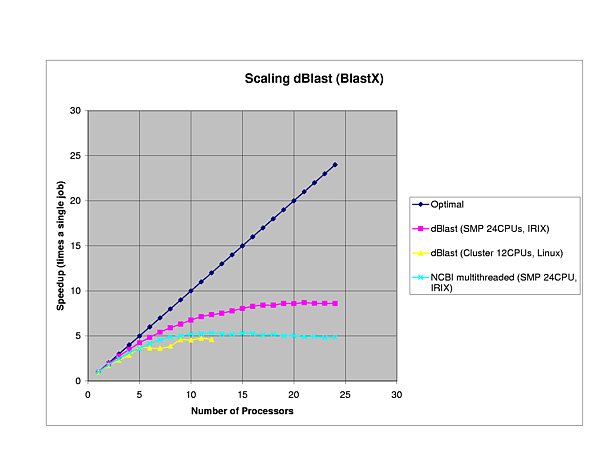
We performed several benchmarks on two platforms: a 24-CPU Silicom Graphics Origin 3800 and a 6 node (dual-CPU, Intel Pentium III-1Ghz) Linux cluster.
Query Sequences:
Protein: PHY1_SYNY3 (Swissprot Acc.Nr.Q55168, 748 AA)
Nucleic: HSRDS (M73531, the first 1140bp)
Databases:
Protein: SwissProt (121.333 sequences, 44.504.923 AA)
Nucleic:EMBL human division (233.004 sequences, 3.725.671.508 bases)

|

|

|

|

|

|
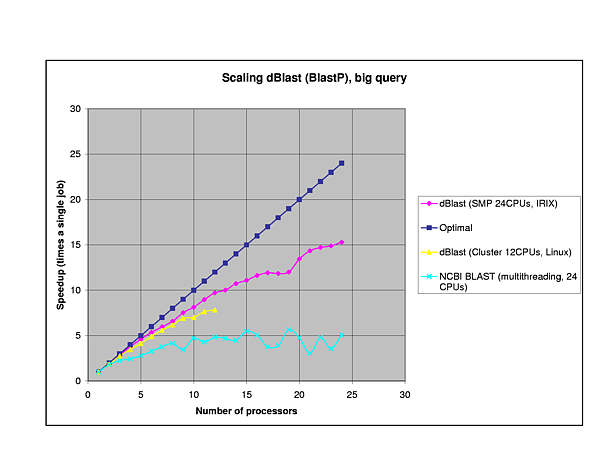
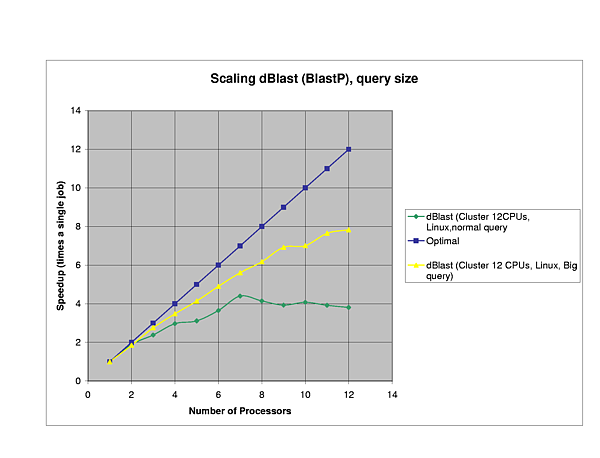
The efficiency of dBlast is there where it is most needed, in the more complex algorithms TBlastX and TBlastN. In general, dBlast is performing better in more complex situations (query/database size). Two examples of the influence of the query size are plotted below.
Query Sequences:
Protein: PHY1_SYNY3 (Swissprot Acc.Nr.Q55168, 748 AA)
NEBU_HUMAN (Swissprot Acc.Nr.P20929, 6669 AA)
Database:
Protein: SwissProt (121.333 sequences, 44.504.923 AA)
|
|
Results obtained by this program can freely be published provided the program and its authors are acknowledged by name. Please use the following reference:
van Driel, Marc A., Hekkelman, Maarten L., and Rodriguez R. (submitted). dBlast - A wrapper to run NCBI BLAST parallel/distributed. submitted


dBlast was written by Marc A. van Driel and Maarten L. Hekkelman.