by Miaomiao Zhou1,3§, Daniel Theunissen2,3, Michiel Wels1,2,3 and Roland Siezen1,2,3.
Abstract
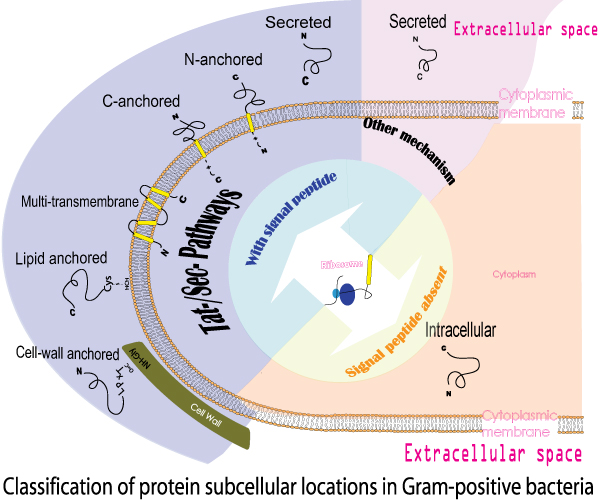 Lactic acid bacteria (LAB) have been broadly used for food fermentations and as inducers of a healthy gut microbiota. Extracellular (extracellular milieu, GO:0005576) and surface-asscociated (anchored to membrane GO:0046658, intrinsic to external side of plasma membrane GO:0031233 and the cell wall, GO: 0005618) proteins play an important role in the related processes ranging from probiotic effects in the gastrointestinal tract to degradation of complex extracellular carbon sources such as found in plant materials. These proteins are secreted by the bacterium and most remained attached to the surface of the cell and as such have a primary role in the communication of a bacterium with its environment [1]. Lactic acid bacteria (LAB) have been broadly used for food fermentations and as inducers of a healthy gut microbiota. Extracellular (extracellular milieu, GO:0005576) and surface-asscociated (anchored to membrane GO:0046658, intrinsic to external side of plasma membrane GO:0031233 and the cell wall, GO: 0005618) proteins play an important role in the related processes ranging from probiotic effects in the gastrointestinal tract to degradation of complex extracellular carbon sources such as found in plant materials. These proteins are secreted by the bacterium and most remained attached to the surface of the cell and as such have a primary role in the communication of a bacterium with its environment [1].
The Lab-Secretome database aims to perform in-depth comparative analysis on the predicted secretomes from sequenced LAB strains. The current version of our database included 26 secretomes (3608 proteins) which have been classified into 462 clusters (LaCOGs),180 strain/species specific proteins (Orphans) and 69 proteins that have only distant homologs in the non-LAB species.
An in-depth comparative analysis of the clustered protein families showed that the absence/presence of various protein families in different LAB species is relevant to known biological niches. In many cases we were able to link genotype to phenotype by combining domain composition and reconstruction of phylogenies. This comparison of secretome proteins of LAB will expand our understanding of the molecular evolution, diversity, function and adaptation of lactic acid bacteria to specific environments. Our methodology is applicable to secretome proteins of all gram-positive bacteria.
§Corresponding author: Miaomiao Zhou
Reference:[1] Kleerebezem M, P Hols, E Bernard, T Rolain, M Zhou, RJ Siezen, P Bron (2010) The extracellular biology of the lactobacilli. FEMS Microbiology Reviews 34, 199-230.
[2] Zhou M, Theunissen D, Wels M, Siezen RJ (2010)LAB-Secretome: A genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of Lactic Acid Bacteria BMC Genomics 2010 Nov 23;11(1):651.
1 Centre for Molecular and Biomolecular Informatics, Radboud University Nijmegen Medical Centre, PO Box 9101, 6500 HB Nijmegen, The Netherlands
2 TI Food and Nutrition, and Kluyver Centre for Genomics of Industrial Fermentation, Wageningen, The Netherlands
3 NIZO Food Research, Ede, The Netherlands





|
